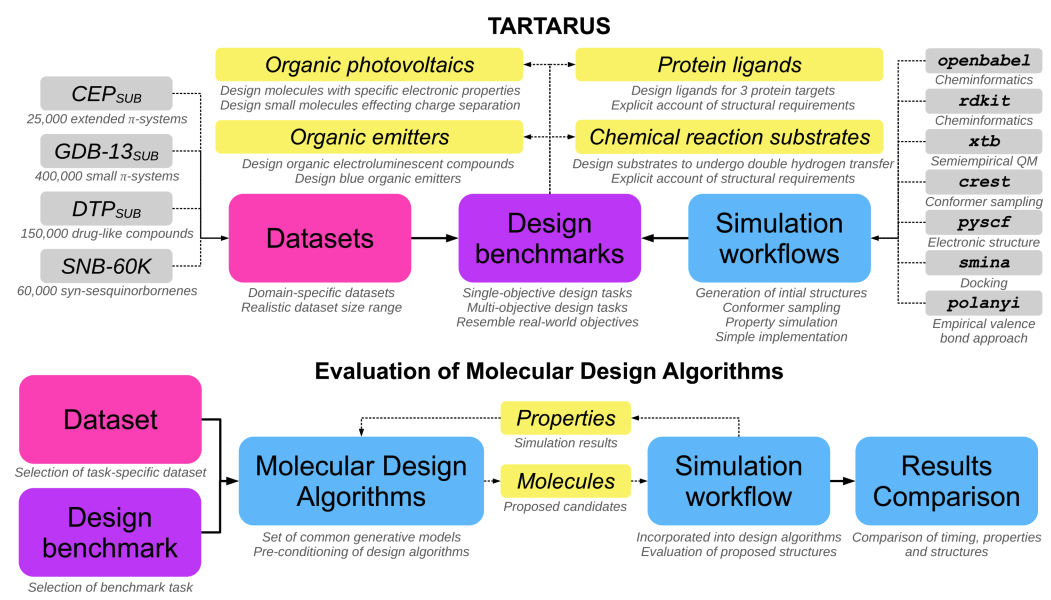
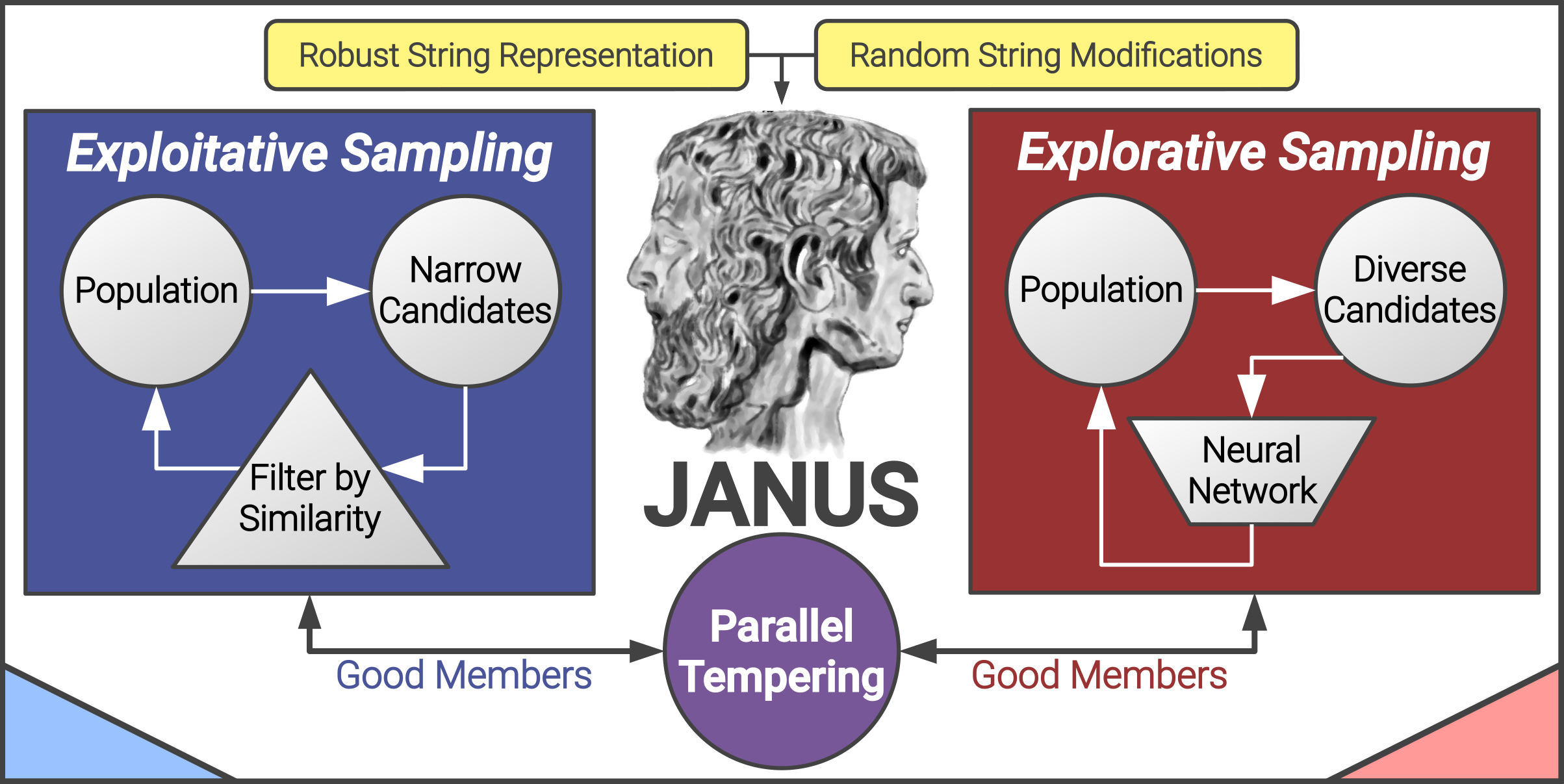
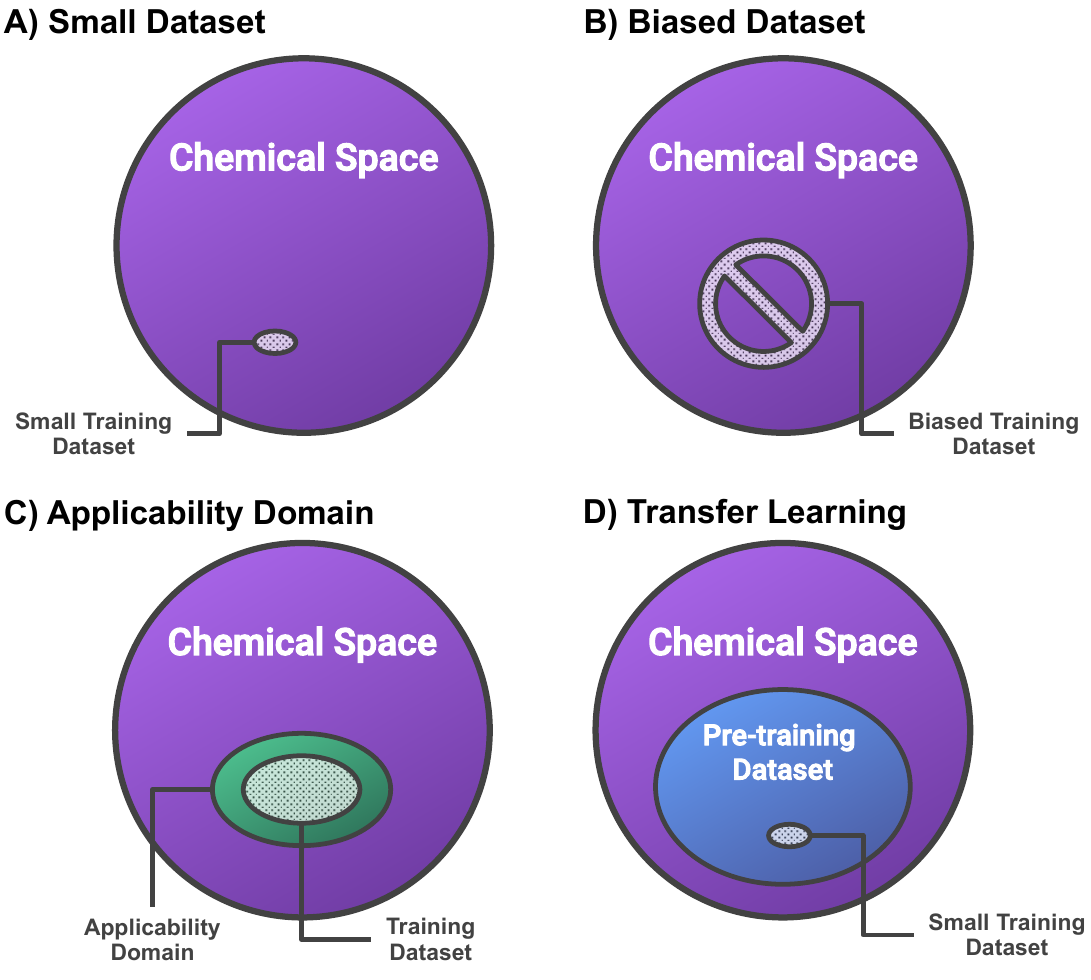
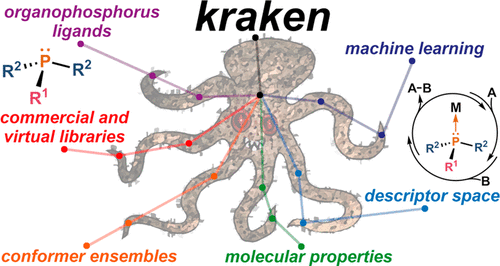
AkshatKumar (Akshat) Nigam
Stanford University · Department of Computer Science & Genetics ·
akshat98@stanford.edu
I am a PhD student at Stanford in the departments of Computer Science & Genetics. I love molecules, both big and small, calculating complex properties and leveraging machine learning tools for estimating those properties. For a detailed list of my publications, please have a look at my Google Scholar